UNITED STATES
SECURITIES AND EXCHANGE COMMISSION
Washington, D.C. 20549
FORM
CURRENT REPORT
Pursuant to Section 13 OR 15(d)
of the Securities Exchange Act of 1934
Date of Report (Date of earliest event reported):
(Exact name of registrant as specified in its charter)
| (State or other jurisdiction of incorporation) |
(Commission File Number) |
(I.R.S. Employer Identification No.) |
(Address of principal executive offices, including zip code)
(Registrant’s telephone number, including area code)
Not Applicable
(Former Name or Former Address, if Changed Since Last Report)
Check the appropriate box below if the Form 8-K filing is intended to simultaneously satisfy the filing obligation of the registrant under any of the following provisions:
| Written communications pursuant to Rule 425 under the Securities Act (17 CFR 230.425) |
| Soliciting material pursuant to Rule 14a-12 under the Exchange Act (17 CFR 240.14a-12) |
| Pre-commencement communications pursuant to Rule 14d-2(b) under the Exchange Act (17 CFR 240.14d-2(b)) |
| Pre-commencement communications pursuant to Rule 13e-4(c) under the Exchange Act (17 CFR 240.13e-4(c)) |
Securities registered pursuant to Section 12(b) of the Act:
| Title of each class |
Trading |
Name of each exchange | ||
Indicate by check mark whether the registrant is an emerging growth company as defined in Rule 405 of the Securities Act of 1933 (§ 230.405 of this chapter) or Rule 12b-2 of the Securities Exchange Act of 1934 (§ 240.12b-2 of this chapter).
Emerging growth company
If an emerging growth company, indicate by check mark if the registrant has elected not to use the extended transition period for complying with any new or revised financial accounting standards provided pursuant to Section 13(a) of the Exchange Act.
Item 7.01. Regulation FD Disclosure
On October 24, 2022, Monte Rosa Therapeutics, Inc. presented at a Key Opinion Leader (KOL) webinar hosted by Cowen and Company, LLC on the topic of MRT-2359. The full KOL webinar presentation is furnished as Exhibit 99.1 to this Current Report on Form 8-K.
The information in this Form 8-K (including Exhibit 99.1) shall not be deemed “filed” for purposes of Section 18 of the Securities Exchange Act of 1934, as amended (the “Exchange Act”), or otherwise subject to the liabilities of that section, nor shall it be deemed incorporated by reference in any filing under the Securities Act of 1933, as amended, or the Exchange Act, except as expressly set forth by specific reference in such a filing.
Item 9.01. Financial Statements and Exhibits
(d) Exhibits
| 99.1 | KOL webinar presentation furnished by Monte Rosa Therapeutics, Inc. on October 24, 2022. | |
| 104 | Cover Page Interactive Data File (embedded within the Inline XBRL document). | |
SIGNATURE
Pursuant to the requirements of the Securities Exchange Act of 1934, the registrant has duly caused this report to be signed on its behalf by the undersigned hereunto duly authorized.
| Monte Rosa Therapeutics, Inc. | ||||||
| Date: October 24, 2022 | By: | /s/ Markus Warmuth | ||||
| Markus Warmuth | ||||||
| President and Chief Executive Officer | ||||||

Exhibit 99.1 MRT-2359 KOL Webinar hosted by Cowen October 24, 2022 Guest Speakers Jordi Rodon Ahnert, M.D., Ph.D., Department of Investigational Cancer Therapeutics, Division of Cancer Medicine at MD Anderson Cancer Center Davide Ruggero, Ph.D., Professor, Department of Urology and Cellular & Molecular Pharmacology at UCSF; Helen Diller Family Endowed Chair in Basic Cancer Research

Forward-Looking Statements These materials include express and implied “forward-looking statements,” including forward-looking statements within the meaning of the Private Securities Litigation Reform Act of 1995. Forward looking statements include all statements that are not historical facts, and in some cases, can be identified by terms such as “may,” “might,” “will,” “could,” “would,” “should,” “expect,” “intend,” “plan,” “objective,” “anticipate,” “believe,” “estimate,” “predict,” “potential,” “continue,” “ongoing,” or the negative of these terms, or other comparable terminology intended to identify statements about the future. Forward-looking statements contained in herein include, but are not limited to, statements about our product development activities, including our expectations around MRT-2359 and the ongoing development of our QuEEN™ platform, and the advancement of our pipeline and the various products therein, our expectations of timing for initiation of our clinical trial for MRT-2359, our expectations of timing for dosing patients in our clinical trial for MRT-2359, our ability to initiate and the timing of initiation of additional lead optimization programs, and our expectations regarding our ability to nominate and the timing of our nominations of additional development candidates. By their nature, these statements are subject to numerous risks and uncertainties, including the impact that the current COVID-19 pandemic will have on our development activities and operations, as well as those risks and uncertainties set forth in our Annual Report on Form 10-K for the fourth quarter and full year ended December 31, 2021 filed, with the US Securities and Exchange Commission on March 29, 2022, and any subsequent filings, including our Quarterly Report on Form 10-Q for the second quarter of 2022 ending on June 30, filed on August 11, 2022, that could cause actual results, performance or achievement to differ materially and adversely from those anticipated or implied in the statements. You should not rely upon forward looking statements as predictions of future events. Although our management believes that the expectations reflected in our statements are reasonable, we cannot guarantee that the future results, performance or events and circumstances described in the forward-looking statements will be achieved or occur. Recipients are cautioned not to place undue reliance on these forward-looking statements, which speak only as of the date such statements are made and should not be construed as statements of fact. We undertake no obligation to publicly update any forward-looking statements, whether as a result of new information, any future presentations or otherwise, except as required by applicable law. Certain information contained in these materials and any statements made orally during any presentation of these materials that relate to the materials or are based on studies, publications, surveys and other data obtained from third-party sources and our own internal estimates and research. While we believe these third-party studies, publications, surveys and other data to be reliable as of the date of these materials, we have not independently verified, and make no representations as to the adequacy, fairness, accuracy or completeness of, any information obtained from third-party sources. In addition, no independent source has evaluated the reasonableness or accuracy of our internal estimates or research and no reliance should be made on any information or statements made in these materials relating to or based on such internal estimates and research. These materials remain the proprietary intellectual property of Monte Rosa Therapeutics and should not be distributed or reproduced in whole or in part without 2 the prior written consent of Monte Rosa Therapeutics.

MYC Transcription Factors are Undruggable Oncogenes MYC family members are amongst the most Cells expressing high MYC are sensitive to MYC CRISPR KO dysregulated oncogenes in human cancer NSCLC – N-MYC SCLC – L-MYC • MYC family: c-MYC, N-MYC, and L-MYC • MYC dysregulation is frequently associated with poor prognosis and unfavorable patient survival • MYC up-regulation dysregulates key cellular processes (e.g. ribosome biogenesis and protein synthesis) • MYC dependency is observed in many cancer types DepMap data, each dot represents a cell line Targeting enhanced translation induced by MYC represents an attractive alternative 3
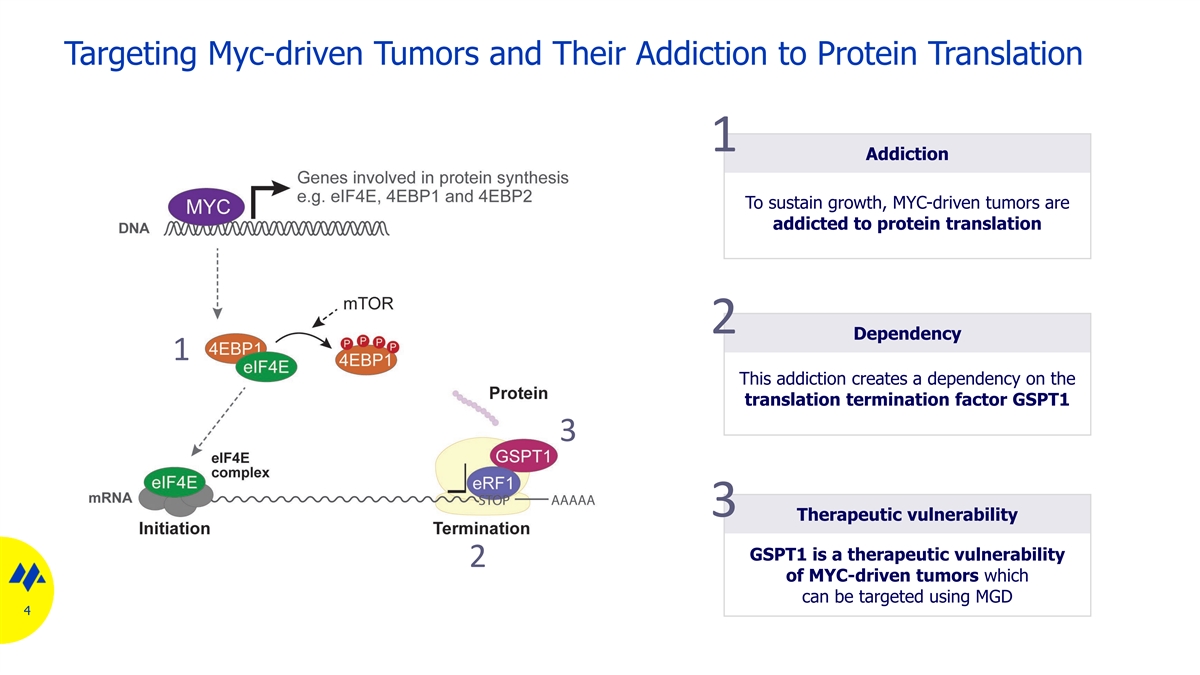
Targeting Myc-driven Tumors and Their Addiction to Protein Translation 1 Addiction To sustain growth, MYC-driven tumors are addicted to protein translation 2 Dependency 1 This addiction creates a dependency on the translation termination factor GSPT1 3 3 Therapeutic vulnerability GSPT1 is a therapeutic vulnerability 2 of MYC-driven tumors which can be targeted using MGD 4

QuEEN™ Discovery Engine Facilitates the Discovery of MRT-2359 MRT-2359 is a potent Proprietary MGD library Rhapsody™ GSPT1 degrader Diverse library, rationally In silico ternary complex GSPT1 designed, using structural insights modelling using proprietary to engage a variety of degrons AI-powered algorithms Degron MRT-2359 CRBN in vitro data CRBN binding, K 113 nM i Ternary complex, EC < 7 nM 50 Degradation, DC 80 nM 50 5

MRT-2359 is a GSPT1-directed MGD with Favorable Drug-like Properties MRT-2359 is orally bioavailable and MRT-2359 is a selective GSPT1-directed MGD has favorable ADMET profile - 0.3 3 30 30 30 Proximity – Turbo ID MRT-2359, µM ADMET profile - - - - + - Bortezomib - - - - - + CYP DDIs > 30 µM MLN-4924 GSPT1 hERG inhibition EC > 30 µM 50 patch clamp IKZF1 Oral bioavailability ~50% IKZF3 all species SALL4 • MRT-2359 is neither an inhibitor, ZFP91 nor an inducer of major CYPs • MRT-2359 doesn’t inhibit hERG CK1a • MRT-2359 is orally bioavailable Protein fold-change; (log ) 2 GAPDH 6hr post treatment in MM1S and Kelly (SALL4) 1hr post treatment 6 p-value

Preferential activity of MRT-2359 in MYC-Driven NSCLC Lines MRT-2359 induces GSPT1 degradation in all cell models, but N-MYC overexpression sensitizes show preferential antiproliferative activity in N-MYC high cell lines NCI-H2023 resistant cells to MRT-2359 GSPT1 degradation Viability Doxycycline-inducible N-MYC model 1.00 100 100 - Dox Wash-out 0.75 75 75 0.50 50 50 0.25 25 25 + Dox 0.00 0 0 Dox - + + 0.00010.001 0.01 0.1 1 10 100 0.0001 0.001 0.01 0.1 1 10 0.0001 0.001 0.01 0.1 1 10 Wash-out - - + MRT-2359,µM MRT-2359,µM MRT-2359,µM N-MYC GAPDH NCI-H2023 Dox-inducible MYC - Dox High N-MYC Low N-MYC NCI-H2023 Dox-inducible MYC + Dox NCI-H1155 NCI-H2023 NCI-H2023 Dox-inducible MYC + Dox wash-out ABC-1 NCI-H441 7 GSPT1 western blot at 6 hr (N-Myc high) and 24 hr (low). 72 hr viability assay (CTG) Incucyte, 96 hr post treatment GSPT1 (relative levels) Viability (%) Area under the curve (% to DMSO)
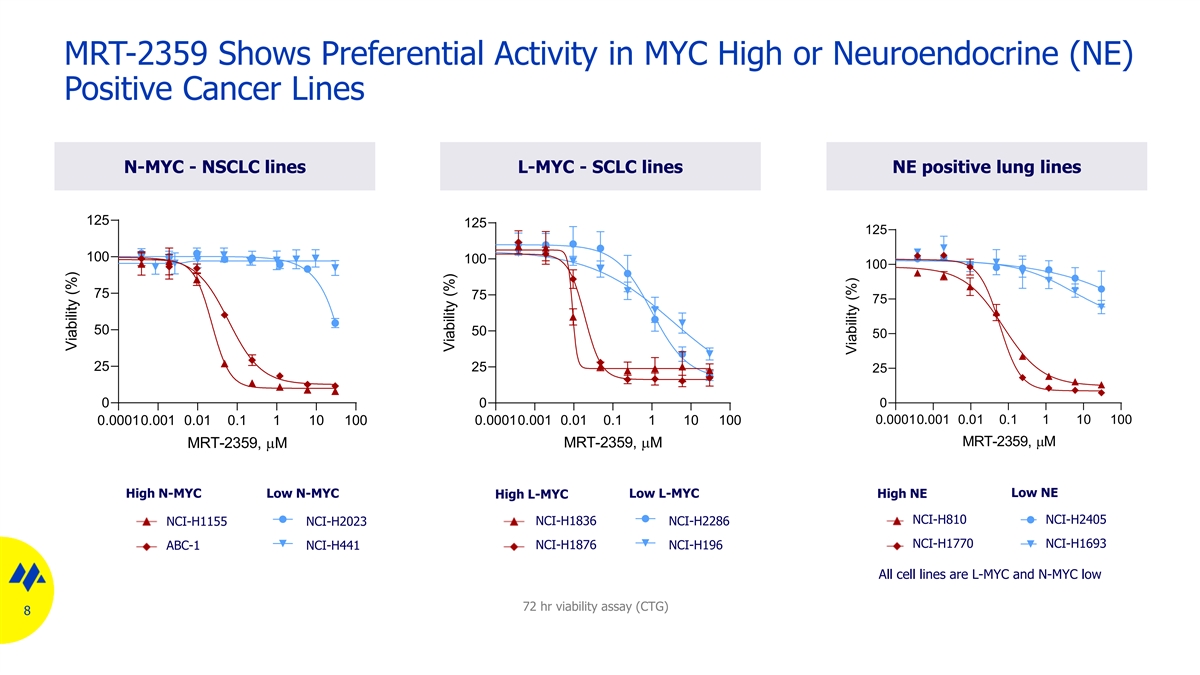
MRT-2359 Shows Preferential Activity in MYC High or Neuroendocrine (NE) Positive Cancer Lines N-MYC - NSCLC lines L-MYC - SCLC lines NE positive lung lines 125 125 125 100 100 100 75 75 75 50 50 50 25 25 25 0 0 0 0.00010.001 0.01 0.1 1 10 100 0.00010.001 0.01 0.1 1 10 100 0.00010.001 0.01 0.1 1 10 100 MRT-2359,µM MRT-2359,µM MRT-2359,µM High N-MYC Low N-MYC Low L-MYC Low NE High L-MYC High NE NCI-H810 NCI-H2405 NCI-H1836 NCI-H1155 NCI-H2023 NCI-H2286 NCI-H1770 NCI-H1693 ABC-1 NCI-H441 NCI-H1876 NCI-H196 All cell lines are L-MYC and N-MYC low 72 hr viability assay (CTG) 8 Viability (%) Viability (%) Viability (%)

MRT-2359 Shows Preferential Activity Compared to “Rapid” GSPT1 Degraders MRT-2359 ¨Rapid¨ GSPT1 degraders lack preferantial actvity in N-MYC high cell lines 100 100 100 75 75 75 50 50 50 25 25 25 0 0 0 0.00010.001 0.01 0.1 1 10 100 0.00010.001 0.01 0.1 1 10 100 0.00010.001 0.01 0.1 1 10 100 MRT-2359,µM CC-885,µM MRT-xxxx (rapid degrader),µM High N-MYC Low N-MYC • Differential activity can be optimized and is a funtion of selectivity and degradation NCI-H1155 NCI-H2023 dynamics ABC-1 NCI-H441 • High selectivity and intermediate fast degradation (6h – vs 1-2h to maximum degradation) lead to greater differential activity 9 72 hr viability assay (CTG) Viability (%) Viability (%) Viability (%)

Translation Initiation/Elongation Inhibitors Do Not Show Preferential Activity in MYC High NSCLC and SCLC Cell Lines MRT-2359 shows preferentially Translation initiation and elongation inhibitors lack differential activity activity in MYC high lung lines MRT-2359 Initiation inhibitor Elongation inhibitor mTOR inhibitor (eIF4Ai, Zotatifin) (Homoharringtonine) (rapamycin) N-MYC L-MYC N-MYC L-MYC N-MYC L-MYC N-MYC L-MYC N-MYC L-MYC N-MYC L-MYC N-MYC L-MYC N-MYC L-MYC low high /NE low high /NE low high /NE low high /NE low high /NE low high /NE low high /NE low high /NE 10 NSCLC SCLC NSCLC SCLC NSCLC SCLC NSCLC SCLC 72 hr viability assay (CTG). EC (nM) 50

MRT-2359 Mechanism of Action

MRT-2359 Mechanism of Action in MYC-driven Tumors Synthetic lethality MRT-2359 impairs protein synthesis in MYC high lines Oncogene addiction MRT-2359 affects MYC and downstream in MYC high lines 12

MRT-2359 Impairs Protein Synthesis in N-MYC High NSCLC Cell Lines MRT-2359 induces ribosome stalling MRT-2359 rapidly and completely abrogates only in N-MYC high cell line protein synthesis only in N-MYC high cell line Peak density GSPT1 protein levels Puromycin incorporation 180 120 DMSO 160 MRT-2359 6 hr 100 MRT-2359 MRT-2359 MRT-2359 48 hr 140 DMSO 6 hr 48 hr 80 120 Low N-MYC 100 60 GSPT1 NCI-H2023 80 40 60 GAPDH 20 40 20 0 0 DMSO MRT-2359 0 10 20 30 40 50 60 70 80 90 Time (min) Peak density GSPT1 protein levels Puromycin incorporation 180 DMSO 120 160 MRT-2359 6 hr 140 100 120 MRT-2359 80 100 High N-MYC 6 hr DMSO 60 80 NCI-H1155 GSPT1 60 40 40 20 20 GAPDH 0 0 0 10 20 30 40 50 60 DMSO MRT-2359 Time (min) 13 Peak density Peak density Puromycin incorporation Puromycin incorporation

MRT-2359 Affects MYC and MYC Pathway in N-MYC High NSCLC Cell Lines MRT-2359 induce GSPT1 degradation leading Degradation of GSPT1 leads to downregulation to N-MYC protein downregulation in NCI-H1155 of N-MYC transcriptional output in NCI-H1155 Time course RNAseq 0 6 hr 24 hr 48hr MRT-2359 (μM) - - - - Low N-MYC GSPT1 NCI-H2023 N-Myc not detected N-MYC Tubulin Time course RNAseq 0 6 hr 24 hr 48hr MRT-2359 (μM) - - - - High N-MYC GSPT1 Transcriptional NCI-H1155 modulation of >200 N-MYC MYC targets genes Tubulin 14 MYC targets gene set score MYC targets gene set score

MRT-2359 and Other Clinical Stage GSPT1 Degrader

MRT-2359 Shows Superior Characteristics Compared to Clinical GSPT1 Degrader Assay MRT-2359 Clinical GSPT1 Degrader Selectivity (TMT Px, WB) GSPT1, GSPT2 GSPT1, GSPT2, SALL4, FIZ1, RNF166, ODC1 CYP DDI > 30 uM CYP2C19 @ 1.5 uM (2B6, 1A2, 2D6,3A4, 2C8,2C9, 2C19) > 30 uM 5.3 uM hERG (patch clamp) a1A > 50% @ 10 uM M1/M2 > 50% @ 10 uM CEREP Caco2 (Efflux Ratio) 9 >100 Route of Administration PO IV Development status Ph I Phase I/Ib Myc high None reported Stratification * Comparison based on internal profiling. Selectivity based on internal data as well as data from DFCI Proteomic data base https://proteomics.fischerlab.org 16 Clinical in vivo in vitro

Superior Activity of MRT-2359 in MYC-driven Cancer Cell Lines Low MYC High MYC Low MYC High MYC Low MYC High MYC Low MYC High MYC Lymphoma Lymphoma NSCLC SCLC NSCLC SCLC 17 * Comparison based on internal profiling MRT-2359 EC (nM) 50 Clinical GSPT1 Degrader EC (nM) 50

Preclinical Anti-tumor Activity of MRT-2359 in MYC-driven Animal Models

MRT-2359 Mouse-trial in NSCLC, SCLC and Lung NE Patient-derived Xenograft Selected 48 models NSCLC Collection of PDX models Models selected across range L-MYC mRNA expression of N-MYC and L-MYC mRNA Log2 (TPM+1) expression levels or NE status were treated with: § Vehicle Selected § MRT-2359 10 mg/kg PO QD 20 models SCLC All models have been characterized by DNA and RNAseq 3 mice for each treatment group L-MYC mRNA expression Log2 (TPM+1) Selected Large cell NE carcinoma 10 models or NE lung cancer 19 NE genes Log FPKM 2 N-MYC mRNA expression N-MYC mRNA expression Models Log2 (TPM+1) Log2 (TPM+1)

MRT-2359 Demonstrates Preferential Anti-tumor Activity in MYC High or Neuroendocrine (NE) Lung Cancer PDXs NSCLC SCLC NE Lung Cancer Large cell NE carcinoma or NE lung cancer L-MYC and N-MYC low L-MYC or N-MYC high N-MYC and L-MYC low 20 MRT-2359 10 mg/kg, PO, QD

High Frequency of L-MYC and N-MYC Expression in NSCLC and SCLC from Real-world Data NSCLC SCLC 3053 samples 188 samples Demographic and Diseases Characteristic • There is no notable difference in the proportion of MYC high expressors across disease staging, gender or racial groups Treatment Outcomes • No statistically significant associations between MYC high status and treatment outcomes L-MYC mRNA expression Log2 (TPM+1) mRNA expression High N-MYC or L-MYC 21 Low N-MYC and L-MYC N-MYC mRNA expression Log2 (TPM+1)

Phase 1/2 Clinical Study

MRT-2359-001 Clinical Study Design Phase 2: Expansion Cohorts Phase 1: Dose Escalation Lung cancer (NSCLC & SCLC), DLBCL, high-grade neuroendocrine tumors, and N-/L-MYC amplified solid tumors MTD or RP2D NSCLC* – high N- or L-MYC expression Dose level X – low N- and L-MYC expression Dose level 4 Backfill SCLC* – high N- or L-MYC expression Dose level 3 Backfill – low N- and L-MYC expression Dose level 2 Backfill Dose level 1 Backfill Solid tumors – N- or L-MYC amplification Backfill slots for additional patients for each dose level * Efficacy guided stratification per N-/L-MYC expression 23
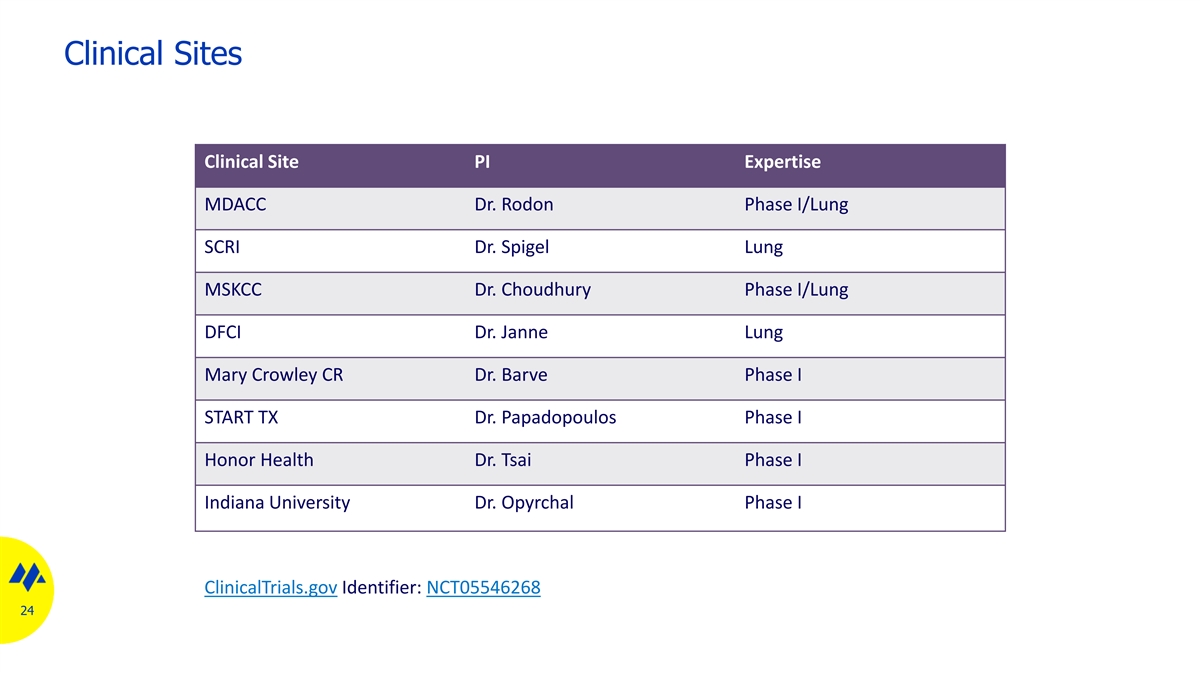
Clinical Sites Clinical Site PI Expertise MDACC Dr. Rodon Phase I/Lung SCRI Dr. Spigel Lung MSKCC Dr. Choudhury Phase I/Lung DFCI Dr. Janne Lung Mary Crowley CR Dr. Barve Phase I START TX Dr. Papadopoulos Phase I Honor Health Dr. Tsai Phase I Indiana University Dr. Opyrchal Phase I ClinicalTrials.gov Identifier: NCT05546268 24

Thank You
